MAT Homeodomain-DNA Structure
Reference:
Li, 1995. Crystal structure of the MATa1/MATalpha2 homeodomain
heterodimer bound to DNA Science 270, 262-269. [Medline
abstract]
Click this button
I. Introduction
The A1/alpha2 - DNA Complex exhibits several properties which
are important in DNA protein interactions:
- Combinatorial binding of proteins to DNA
- Protein-protein interactions
- DNA bending induced by protein binding
- H-bonding and water mediated H-bonding
a1 and alpha2
regulate yeast mating type loci:
- diploid cells: a1/alpha2
formed - shuts off haploid-specific genes
- haploid (alpha) cells: MCM1/alpha2
formed - shuts off a-specific genes
So MCM1 and a1 can change
binding specificity of alpha2.
All are homeodomain proteins- originally discovered in Drosophila
homeotic genes, a gene family with a conserved 60 aa sequence
near the C-terminus (the homeodomain). Many have been shown
to regulate development in various organisms. They belong to the
"helix-turn-helix" class of DNA binding proteins and
often act in combination with other homodomain proteins. The "helix-turn-helix"
proteins have an alpha helix which fits into the major groove
of the DNA, backed by 2 alpha helices which lay across the groove.

Both a1 and alpha2
bind DNA by themselves, but with low affinity and specificity.
Both binding specificity and affinity are increased 105-fold
when they bind as a homeodimer. Binding is dependent on presence
of both binding sites at proper spacing.
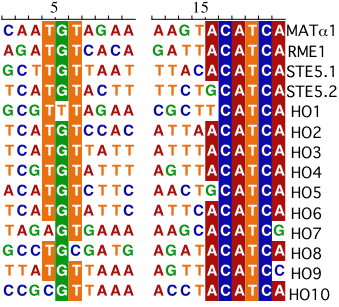
The sequences above are sites to which the a1/alpha2 heterodimer binds in vivo.
The boxed regions are conserved in at least 75% of the yeast binding
sites for MATa1/alpha2. Note that the conserved regions are spaced
the same distance apart at all sites.
II. The complex
The crystal structure of the a1/alpha2 heterodimer complexed with
DNA is shown at left, similar to Fig1a of the reference. To make
the coloring match the refernce and the rest of the figures of
this tutorial, click this button
III. Protein-protein interaction
The a1/alpha2
protein-protein interaction can be seen by clicking here
. A hydrophobic patch of the a1
homeodomain interacts with hydrophobic residues of the alpha2 C-terminal tail. Note the way
the hyrophobic amino acids interdigitate between the two proteinThis
interaction resembles the Oct1/VP16 interaction; these proteins
share homology with a1/alpha2
in this region (see alignment above). Mutation of the hydrophobic
residues to A results in loss of a1/alpha2 binding activity, based on
an in vivo reporter gene assay. H-bonds, shown as
dotted lines here , also
contribute to this interaction.
IV. Protein-DNA contacts
A diagram of the DNA-protein contacts is shown below. H-bonds
are indicated by arrows, and the proteins are color coded as above.
Water molecules involved in H-bonds are shown as blue "w"
s. Conserved nucleotides are shown in green.
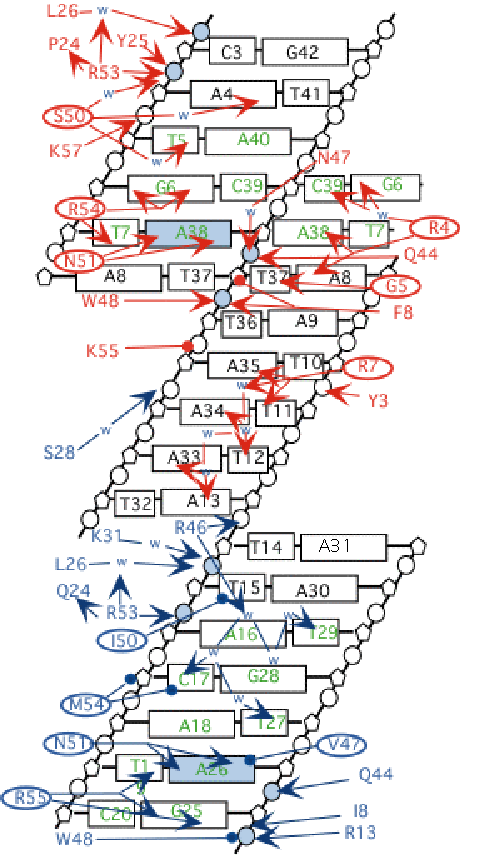
This button colors the structure as
in fig 6B, showing interactions of a1
with the major groove of the DNA. N51 forms 2 H-bonds with A26
of the DNA. This interaction is conserved in all homeodomain proteins,
including alpha2 (below).
Also conserved, but not shown, is an H-bond between R53 and the
phosphate of A4. R55 makes 2 H-bonds with G25, an interaction
similar to the N51-A26 interaction. Many other H-bonds are made
through ordered water molecules. Since water can act as both an
H donor and acceptor, these types of interactions are difficult
to predict in the absence of crystallographic data. Red-Blue stereo picture of this view.
This button Shows the interactions
of alpha2 with the major groove,
as in fig. 6D. N51 of alpha2
makes the conserved interaction with an adenosine residue, in
this case A38. R54 makes H-bonds with G6 and to N51 of alpha2 itself. S50 makes 2 water-mediated
H-bonds, one each to A4 and T5.
This button shows the interactions
alpha2 makes with the minor
groove. R4 has several potential H-bond partners. The epsilon
N can pair with N3 of A8 or A38. The terminal amine can make water
mediated bonds with G6 and C39. It is also close enough to make
a direct bond with O2 of C39. The backbone NH of glycine5 donates
an H-bond to T37, and R7 donates an Hbond to A35 and T11. Red-Blue stereo
picture of this view.
 Back
to intro to DNA-RNA structure.
Back
to intro to DNA-RNA structure.
 This section Still Under Construction
This section Still Under Construction
Comments or Suggestions to:Jim
Nolan at James.M.Nolan@tulane.edu


