U1A RNA-protein complex
7783201
Reference: Oubridge, C., Ito, N., Evans,
P.R., Teo, C.-H., and Nagai, K. (1994). Crystal structure at 1.92A
resolution of the RNA-binding domain of the U1A spliceosomal protein
complexed with an RNA hairpin. Nature. 372, 432-438. [Medline
abstract]
Introduction
The U1A signal recognition particle (U1snRNP) is responsible
for selection of 5' splice sites in intron splicing. The U1A protein
binds to Domain B of the U1snRNA in the U1 snRNP. The U1A protein
is a member of the RNP (or RRM) family of proteins, by virtue
of the presence of the characteristic RNP motif in the protein
sequence. The RNP domain is comprised of the RNP1
and RNP2 motifs. Other members
of this family include other snRNP proteins, hnRNP proteins and
poly A binding proteins.
Overview
To color-code the crystal structure at left, click here <
Regions of the RNA which interact with protein are shown in
heavy frames on the 3D structure and are boxed in the secondary
structure below. Unimportant regions are shown as light, white
wireframe. 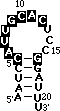
The binding strategy for this interaction is somewhat surprising:
the loop residues are splayed out in an extended conformation
(in contrast to the anticodon loop of tRNA, for example. In the absence of the protein,
the RNA loop appears unstructured, the loop structure seen here
is found only in the presence of the protein, which locks it into
place by an induced-fit mechanism. Residues in the loop which
do not bind protein remain largely unstructured; they vary in
conformation depending upon their location in the crystal lattice.
The protein loop between RNP1
and sheet 2 projects through
the RNA loop. This region varies in sequence between different
members of the RNP family and may account for some of the differences
in specificity between different RNP proteins. This loop region
plus the RNP1 and RNP2 regions make up the bulk of the
contact sites for the RNA. The protein loop is also less structured
in the absence of the RNA, so the induced-fit mechanism is mutual
for both the RNA and protein.
Hydrogen bonds and stacking interactions
<
<
<
<
<
 Back
to intro to DNA and RNA structure.
Back
to intro to DNA and RNA structure.
Comments or Suggestions to:Jim
Nolan
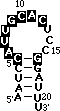
![]() Back
to intro to DNA and RNA structure.
Back
to intro to DNA and RNA structure.