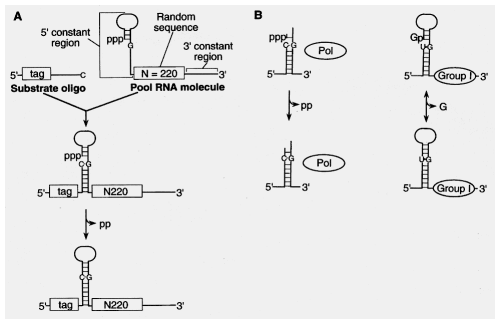
Take advantage of having active groups and genetic info on same molecule.
Gold and Tuerk (1990) Science 249,505-510 (SELEX)
Ellington and Szostak (1990) Nature 346, 818-822. (in vitro selection)
Start with Synthesized oligonucleotide containing random sequence:
need way to select for molecules you want and get rid of those
you don't:
protein binding (aptamers)
column binding " "
self-cleavers or -ligaters (--> change in gel mobility)
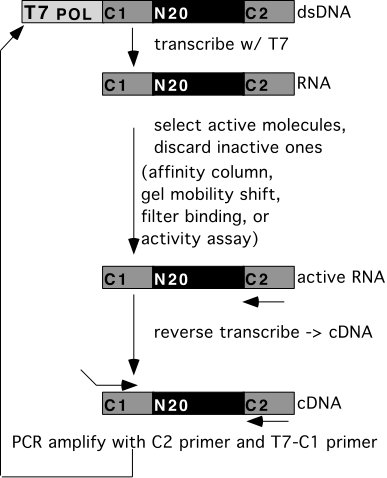
Transcribe RNA
Select active molecules
PCR amplify active molecules (reverse transcribe, then PCR)
repeat many cycles
--> collection of molecules selected to do what you want.
longer random sequence more likely to have wanted functional
groups
but as RNA gets longer, takes more sequence to represent all possible
sequences.
random sequence # possible sequences 20-mer 4e20 =1.1X10e12 =1.8 pmoles = 12 ng 30-mer 4e30 =1.1X10e18 =1.8 umoles = 18 mg 40-mer 4e40 =1.2X10e24 =2 moles! = 26 kg! 165-mer 4e165 =2.2X10e99
6 orders of magnitude/10 additional nts.
Characterization of RNA ligase activities selected by Bartel and Szostak.
Strategy for selection

selected for RNAs that could ligate a tagged oligo to their own 5' end.
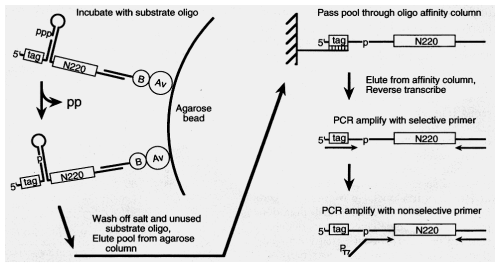
10e15 seqs: wound up using 1.6 mg of RNA in first round.
Used high monovalent and Mg to favor formation of intramolecular structure, but resulted in ppt. (from intermolecular assoc.)
so immobilized RNA on column before adding Mg
--> only intramolecular structures.
Ligation: Cycled temp. to sample different conformations
16hr reaction.
Elute
Select for attachment of substrate oligo.
Used different tag sequences in case of chance affinity of "enzyme" for tag. or resin. (BIG potential problem in selection schemes)
Elute.
Selective PCR (only ligated sequences amplified)
Reconstructive PCR -- get rid of substrate seqs. add back T7 promoter
Transcribe --> reconstructed RNA enzyme ready
to react in next round.
Mutagenic PCR in middle rounds. Allows suboptimal RNAs to evolve into more efficient ones
Reduce reaction time, Mg --> more stringent selection
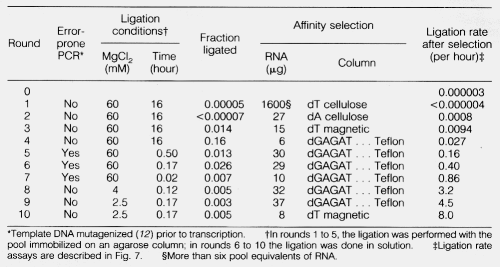
Eckland paper
Characterize RNA and show they can act as true enzyme.
Deletion analysis to identify minimal catalytic domain
Phylogenetic analysis to determine structure of classI ligase:
Resynthesized classI sequence with "doped" oligonucleotides (chemically synthesized so that oligos have small percentage of wrong nucleotides at each base.
In this case for any DNA template for RNA synthesis, 80% of the nucleotides were correct, but 20% would have instead one of the other nucleotides substituted instead
--> lots of inactive mutants, but many active ones.Select for activity and sequence individual active RNAs
Align sequences and analyze or conserved regions and covariations

Biochemical Characterization of enzyme activity
Reconfigure RNA into enzyme and substrate RNAs
run products on gel to characterize:
b1-10t (classI) enzyme looked like the product was migrating faster than the rest
? different linkage formed by ligation?analyzed products by digestion and TLC, revealed that only class I enzyme was forming 3'-5' linkages found in cellular RNA and DNA polymers. Others were 2'-5' links. (This means the reverse transcriptase used to amplify RNA was able to use 2'-5' linkage RNAs as template.
The class I ligase has been used as a starting point for selection of RNA polymerase activity that can faithfully synthesize copies of RNA templates.
Secondary structure of other RNA ligases (2'-5')
use Mn and altered dNTP concs. to increase mutation rate of
PCR
So molecule not present initially may arise from earlier sequence
with lower activity
"In vitro evolution"
Begin with RNA that performs similar function to that desired.
Randomize only part of the molecule likely to be critical for function
--> optimized version of RNA based on existing prototype
also shows variability of less important parts
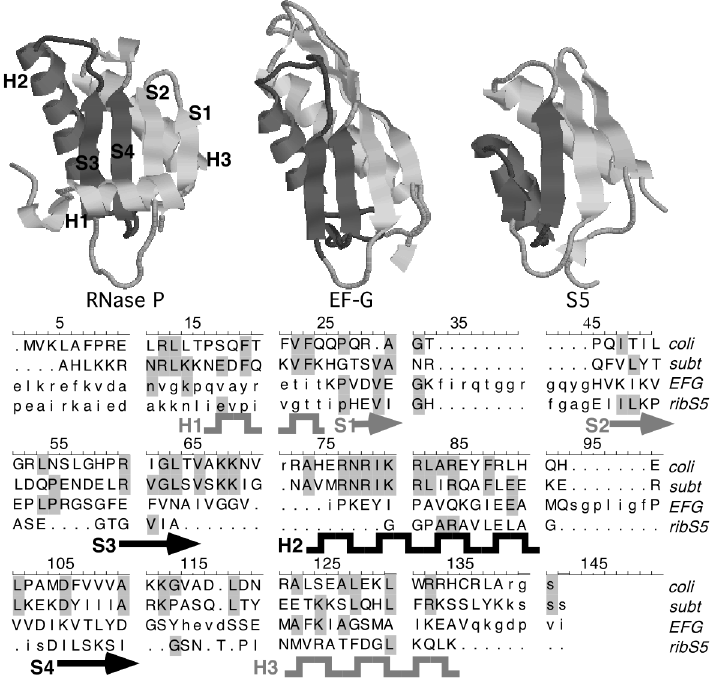
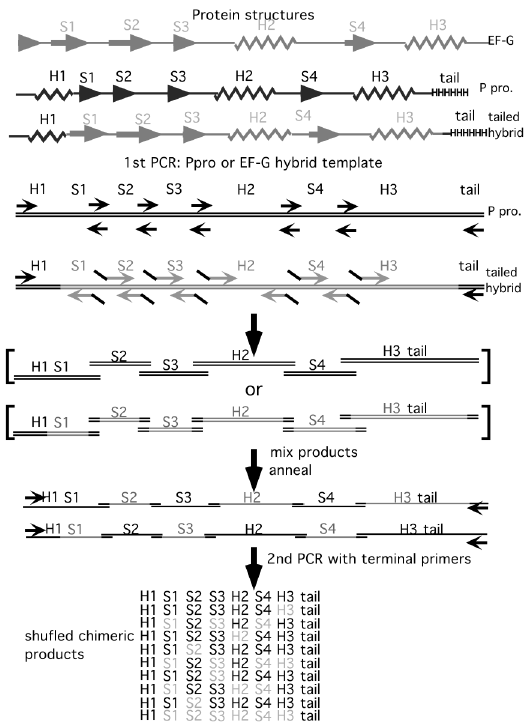
RNAseP protein similar in structure to EF-G.
Use PCR to make chimeric proteins: