To identify important databases for biomedical research
To explain methods for interfacing with databases effectively
Discussion of papers and techniques that utilize bioinformatic and genomic data
There is no required text. Here are a couple of books that I have found helpful:
Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins 2nd Edition by Andreas D. Baxevanis (Editor) – a good general overviewBioinformatics: Sequence and Genome Analysis by David W. Mount – more intense explanations of algorithms
Beginning Perl for Bioinformatics by James Tisdall - good introduction to writing your own programs for use in bioinformatics. Does not assume extensive computer knowledge.
Bioinformatics is defined as:
The use of computers in solving information problems in the life sciences, mainly, it involves the creation of extensive electronic databases on genomes, protein sequences, etc. Secondarily, it involves techniques such as the three-dimensional modeling of biomolecules and biologic systems.by the Online Medical Dictionary.
Since computers, and usually, the
internet, are so heavily involved in the use of bioinformatics, a brief introduction
to how the internet itself works may be beneficial. Much of this info
was obtained from UNH InterOperability
Lab and PC
Lube & Tune.
Lets start by clicking on a web
page link using your internet browser:
for example:
The most important parts of this process are identifying computers named "www.ncbi.nlm.nih.gov" and "me" and negotiating the transfer.

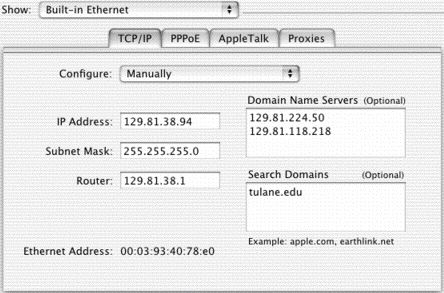
in this case, my IP address is 129.81.38.94.
All Tulane (tulane.edu) computers have addresses beginning with 129.81. The network within Tulane is subdivided into smaller networks (Subnets) interconnected by routers. My computer can connect with any computer with address 129.81.38.### without going through the router, to reach the outside world I need to use the router gateway.
First of all, how do we find www.ncbi.nlm.nih.gov? It doesn't look much like my address. This is accomplished by Domain Name Servers (DNS), computers which keep lists of IP address numbers and corresponding names like "www.tulane.edu," which are easier to remember.
Each institution is responsible for listing all the computers within its domain and the corresponding name, if it has one. The DNS here can query other DNS to see if they have a "www.ncbi.nlm.nih.gov" and if so, what its real number is so we can contact it.
In this case the local DNS is 129.81.224.50 (ns1.tcs.tulane.edu). You may get a "domain name server error" when you can't get through on the network. This could mean that the DNS is down, in which case you might be able to get through to your destination if you know the IP number. But usually this means the connection between you and the network is down, and the first place your computer checks is the DNS.
1 129.81.133.1 (129.81.133.1)2 tidewater-et-4-1.net.tulane.edu (129.81.255.93)3 newsouth-atm-1-0-0.net.tulane.edu (129.81.255.70)4 abilene-houston-pos-oc3.tis.tulane.edu (129.81.255.2)5 atla-hstn.abilene.ucaid.edu (198.32.8.34) University Corporation for Advanced Internet Development6 wash-atla.abilene.ucaid.edu (198.32.8.66)7 wash-abilene-oc48.maxgigapop.net (206.196.177.1) Mid-Atlantic Crossroads (MAX)8 clpk-so3-1-0.maxgigapop.net (206.196.178.46)9 wash-nlm.maxgigapop.net (206.196.177.34)10 130.14.38.185 (130.14.38.185)11 micasaweb.nlm.nih.gov (130.14.22.106)
There is the possibility that unscrupulous people may pretend to be other computers and intercept private data, like credit card numbers. This is why some transfers use secure, encrypted transfers (https instead of http) which prevent others from deciphering what is being sent.
Once the file is sent, you browser determines what kind of file it is (picture, text, or html text file with instuctions for downloading other files embedded in it) and displays the file. The server can tell your computer what kind of file it is sending, like an audio file or spreadsheet, which might be used by another program on your computer.
Both contain virtually all known sequences, including complete genomes
Mostly translated coding sequences from the DNA databaseImportant file formats for both protein and DNA databases are:
GenBank: protein example - DNA example
Currently there are:
more than 100 complete Bacterial genomes
15 complete Archeael genomes
18 complete Eukaryal genomes, including Human
and hundreds of viral genomes
Last year there were:
53 complete Bacterial genomes
11 complete Archeael genomes
10 complete Eukaryal genomes
and hundreds of viral genomes
Thousands of Titles and abstracts from medically relevant journals dating back to the 1960's. Some older citations also available. Powerful searching capabilities essential for identifying articles of interest. Similar databases available for other disciplines (i.e. agricultural)
This page is condensed from the NCBI
PubMed Tutorial Pages . You may find the full tutorial quite useful.
When you enter search terms on the main PubMed
search page, the PubMed server processes your request to attempt to identify
what type of search you are attempting: are you looking up an author name, journal
title, subject area, or phrase from the article abstract? It accomplishes
this by filtering your search terms through successive lists to identify the
types of terms you provide and use them effectively. This process is
called:
Automatic Term Mapping
PubMed compares
your search terms against several lists of search terms to determine what you
are looking for. It checks four lists in order and stops looking once
it finds a match:
The MeSH Translation Table contains:
The Journals Translation Table contains:MeSH terms and Subheadings (searching synonyms for MeSH terms) Chemical Names of Substances
Since MESH terms are searched before Journal Titles, if you want to look up a Journal whose name is also a MESH term, like RNA or Cell, the search will stop with the MESH term and the search for your journal will not be done.Full journal titles MEDLINE title abbreviations International Standard Serial Numbers (ISSN)
The Phrase List contains several hundred thousand phrases generated from:
These are frequently used phrases that are not a part of the MeSH translation tableMeSH Unified Medical Language System (UMLS) Chemical Names of Substances
Author Searching
The format for author searching is last name plus
initials.
PubMed will automatically truncate the author's
name to account for varying initials.
If the term is not found, PubMed will then search the individual words in All Fields.
You can also try putting a phrase in double quotes if the results returned are not what you expected. This will force PubMed to look for the words as a phrase, but it bypasses the Automatic Term Mapping, so you might want to try doing some searches both with and without double quotes.
Truncation
You can truncate a word with the asterisk (*) wildcard This will causes PubMed to return all matches that begin with the truncated string of text. (e.g. enzym* will match enzyme, enzymes, enzymology, enzymatic, etc.) Truncation also turns off Automatic Term Mapping, so the results will be different than nontruncated searches.
Stopwords
PubMed also refers to a list of commonly found words that are referred to as "stopwords ." these are very common words which would match almost every citation and so they are skipped.
The list of stopwords is from PubMed's Help Page.
Stopwords
a did it perhaps these about do its quite they again does itself rather this all done just really those almost due kg regarding through also during km seem thus although each made seen to always either mainly several upon among enough make should use an especially may show used and etc. mg showed using another for might shown various any found ml shows very are from mm significantly was as further most since we at had mostly so were be has must sum what because have nearly such when been having neither than which before here no that while being how nor the with between however obtained their within both I of theirs without but if often them would by in on then can into our there could is overall therefore
Operators
You can use Boolean operators (AND, OR, NOT) to direct your search. These must be entered in UPPERCASE. Operators are processed left-to-right unless you use parentheses to specify the order.
Once you click the "Go" button. Your search is performed and the first 20 hits are displayed in a Summary format:
Author name(s): Title of the article: Brackets indicate a title translated from a foreign language.Source: a brief journal citation. Identification number: A PubMed Unique Identifier (PMID) is included on each record. Links: Includes links to Related Articles and databases, when available.

You can easily scan this first page of citations and see how many of them are really related to what you were trying to find. Though only the first 20 citations are displayed by default (in reverse chronological order) you can see how many total articles matched your search. If you got a surprisingly small or large number of hits, or if there seem to be a high percentage of extraneous hits, you might want to click on the "Details" button in the upper gray box.
Details Button
Clicking Details displays:

Limits ButtonThe PubMed query box shows exactly how PubMed performed your search using the Automatic Term Mapping. It may have found a synonym in the MeSH headings and used that instead of one of your original terms. You can edit the search used and run the edited search by clicking "Search". If the search worked really well, you can save it as a web link by clicking "URL" This formats your search as a URL link your web browser can save as a bookmark to repeat the search at a later date. You can also use the "Cubby" system described below. The "Result "section shows how many hits you got, and links you back to your hits. The translations section describes how each term of your search was interpreted. The database is PubMed, and The User Query is what you typed in to begin with.

Preview/Index ButtonYou can select Publication types (like reviews) from another menu. You can limit searches to specific dates or trials involving subjects in specific age groups, gender, or human/non-human. You can require that hits have Abstracts, though some reviews do not have abstracts, nor do articles indexed before 1975.
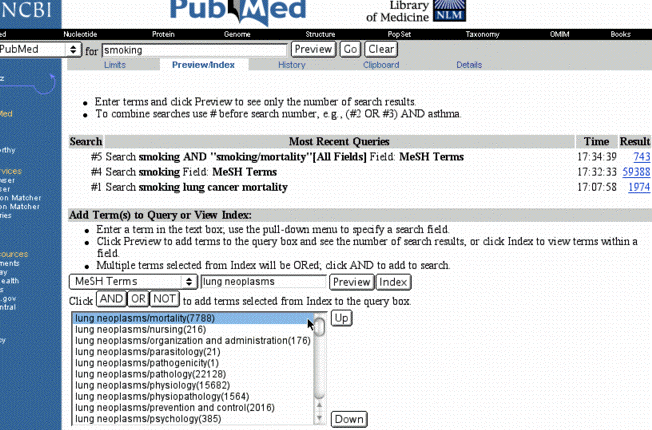
You can have even more control over limits by using
the Preview/Index Feature. You can add search terms by limiting to specific
fields, but you can preview the number of results by clicking on the preview
button.
ResultsBy clicking on index, you can also look up search terms in the index (for example the index of MeSH terms). Items can be added to the search window using the AND, OR, or NOT buttons. Different searches can be combined using their Query number found in the Preview/Index page, a more extensive list is found on the History page. (ex, #4 AND #5). Note that these query numbers disappear after 1 hour of inactivity, so you can't use yesterday's Query number tomorrow and get the same result. You also cannot use these numbers to save your results as a URL in the details window, but you can manually cut and paste the query lines together to save them.
Now that you have constructed the perfect search, you can select the perfect format for displaying results. The default is 20 summary results, but you can choose another format: Other available formats for citation display can be chosen by selecting from the list of choices listed under "Summary":
Brief format includes:Abstract format provides the summary information in addition to:
- First author
- First thirty characters of the title.
- PMID #
- Links
Citation format is similar to abstract, but also includes:
- First Author affiliation
- Abstract, if one is present.
- Links to full-text of the article at provider's Web site, if available.
- Links to Related Articles, Books, LinkOut, and databases.
MEDLINE format is a text file with identifying letters before each field. It is most useful for importing into bibliography programs like EndNote and ProCite.
- MeSH terms.
- Chemical Names of Substances, if any are present.
- Grant numbers, if any are present.
Selecting Citations and Display Format
You can select a subset of the hits to display by clicking the box before each item. If you don't click any boxes, then all are displayed.
Add to Clipboard
- Or you can click on the individual links to see the abstract format for a given citation.
You can select individual citations to save in a clipboard on the server. This is not the clipboard on your computer. After selecting items by clicking their checkbox, click on the "Add to clipboard" link.
Save Button
- The color of item numbers of the hits changes when added to the clipboard.
- If you did not click any boxes, the entire search gets loaded to the clipboard (up to the limit of 500 hits).
- You can view the clipboard by clicking the "Clipboard" link in the features bar. The Clipboard disappears after one hour of inactivity.
You can save citations to a file on your computer by clicking the "Save" link. There is a limit of 10,000 hits. To save selected citations, pick a display format and press "Save". You will be prompted for where to save the downloaded file.
Text Button
You can have the selected items displayed as plain text by clicking the "Text" button. This may be useful for printing if your browser doesn't print the hypertext files well.
Cubby
If you set up a "Cubby", you can save your favorite searches indefinitely on the PubMed server. You have to get a username and password. You can then save your search and rerun it at a later date. Or you can run the search for new articles published since the last time you searched.
LinkOut Preferences
The LinkOut service enables publishers, libraries, biological databases, sequence centers, and other Web resources to display links to their sites on records in PubMed.
You can use Cubby to set which links are displayed by
When you are logged into Cubby, PubMed displays LinkOut providers according to your preferences.
- Adding icons to the Abstract and Citation formats
- Hiding providers from the LinkOut format
Related Articles - Compares words from the title, abstract, and MeSH headings to identify articles similar to the selected article.Related Articles
NCBI Databases
These are the NCBI databases that may be linked to from individual PubMed citations:
- Protein: Protein sequences from SWISSPROT, Protein Information Resource (PIR), Protein Research Foundation (PRF), Protein Data Bank (PDB), and translated protein sequences from the DNA sequences database.
- Nucleotide: DNA sequences from GenBank , European Molecular Biology Laboratory (EMBL), and DNA Data Bank of Japan (DDBJ).
- PopSet: Sequences submitted as a set from a population studies.
- Structure: experimentally-determined, three-dimensional structures.
- Genome: Records and graphic displays of genomes.
- Taxonomy: Index of organisms represented in the sequence databases.
- OMIM: A catalog of human genes and genetic disorders.
- Books provides links to terms described in selected molecular biology textbooks.