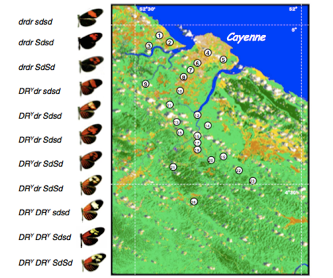
Naturally occurring hybrid zones have been studied for decades because inferences can be drawn from observable patterns of genetic variation to characterize underlying ecological and evolutionary phenomena such as selection across environmental gradients, frequency-dependent selection against hybrids, and Wright’s shifting balance theory. The dynamics of hybrid zones have also been studied to evaluate the stability of range boundaries.
Members of the lab have carried out research on the stability and formation of butterfly hybrid zones in Central and South America. This research continues the long standing (100+ years) tradition of utilizing tropical butterflies (such as Heliconius erato) as model species for studying ecological and evolutionary phenomena. By integrating traditional and molecular genetic approaches with GIS analysis of satellite imagery, it has been possible to demonstrate that concordant hybrid zones at parapatric range boundaries idiosyncratically shift in response to regional land cover change such as deforestation. Unlike studies of shifting range boundaries that focus almost entirely on distributional data, this approach facilitates understanding of how genetic and ecological processes interact to alter species distributions.