Research Description
Our laboratory, Multiscale Bioimaging and Bioinformatics (MBB), has extensive experiences and expertise in biomedical imaging and bioinformatics at both the Biomedical Engineering department, and Biostatistics and Bioinformatics Department at both School of Sciences and Engineering and School of Public Health and Tropical Medicine. In addition, we are part of Tulane Center for Bioinformatics and Genomics (TBG), Tulane Cancer Center (TCC), Computer Science and Neurosciences Program. Our research group has been playing an active role in facilitating the collaboration between biomedical engineering and other departments in the campus.
The following is an outline of our ongoing research projects, which span mathematics, computer sciences, statistics, biomedical engineering and health informatics.
1.Wavelets and sparse representations based signal processing
Given our multi-disciplinary background in mathematical sciences, signal and image processing, biomedical imaging and bioinformatics, the representation and analysis of biomedical imaging and genomic data have been our long interest. We first proposed the B-spline scale-space theory as an alternative to the widely used Gaussian scale-space approach in the area of computer vision and image processing research. The theory we proposed has several advantages over traditional methods. We also develop a differential wavelets, which can have faster computational efficiency, which have found widespread applications in signal and image processing.
a. Yu-Ping Wang, S. L. Lee, Scale-space derived from B-splines, IEEE Trans. Pattern Analysis and Machine Intelligence, vol. 20, no. 10, Oct. 1998, pp.1050-1065.
b. Yu-Ping Wang, S. L. Lee, K. Torachi, Multiscale curvature based shape representation using B-spline wavelets, IEEE Trans. Image Processing, vol. 8, no 11, 1999. pp. 1586-1592.
c. Yu-Ping Wang, Image representations using multiscale differential operators, IEEE Trans. Image Processing, vol. 8, no. 12, 1999, pp. 1757-1771.
d. Yu-Ping Wang, Ruibin Qu. Fast implementation of scale-space by interpolatoy subdivision scheme, IEEE Trans. Pattern Analysis and Machine Intelligence, vol. 21, no. 9, 1999 pp.1050-1065.
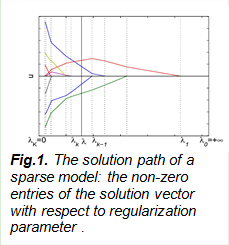
Our recent effort has been the development of sparse representation models, where we propose a theory on an easy condition to recover the sparse signals. A number of papers are currently under review.
e. J. Duan, C. Soussen, D. Brie, J. Idier and Y.-P. Wang, On LARS/homotopy equivalence conditions for over-determined LASSO, IEEE Signal Processing Letters, 19(12), 2012.
The representation and analysis of multi-scale and multi-modal biological data sets is ubiquitous but challenging in many disciplines. We have been working on sparse data representation method, a hot topic in both signal processing, statistics and applied mathematics. The field is more exciting within the context of big data science and technique, which are highly promoted by NIH and NSF, as demonstrated by several program announcements recently. This is especially the case with multi-omics data, where huge amounts of genomic data of different nature, format, organization and structure are produced at different genomic platforms and at multiple scales. These data sets provide rich and complementary information; how to represent and integrate these muli-omics data for systems biology study has become a promising and emerging research topic. We continue this line of research and apply to both imaging and genomic data analysis.
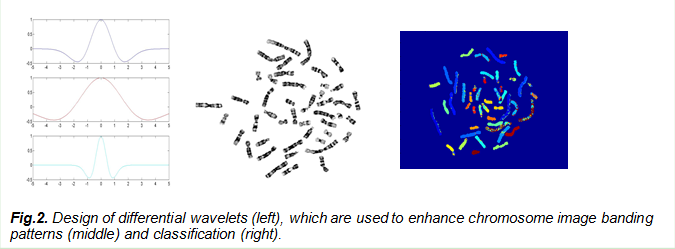
2.Microscopic image processing and industrial software development
The PI, Dr. Wang, has many years of industrial experiences and made contributions to commercializing cytogenetic imaging analysis products. He has worked in bio-industry on various genomic imaging techniques including chromosome G-banding, FISH/M-FISH and microarray imaging under a number of NIH Small Business Innovation Research (SBIR) awards in collaboration with Texas Medical Center in Houston. He was the key team player in the development of PowerGeneTM chromosome karyotyping system and PowerProbeTM FISH system that are now widely used in the market.
a. Yu-Ping Wang, Q. Wu, Ken Castleman, and Z. Xiong, Chromosome Image Enhancement Using Multiscale Differential Operators, IEEE Trans. Medical Imaging, vol. 22, no.5, May, 2003.
b. Z. Liu, Z. Xiong, Q. Wu, Y. Wang, and K. Castleman, Cascaded differential and wavelet compression of chromosome images, IEEE Trans. on Biomedical Engineering, vol. 49, no. 4, 2002.
c. Yu-Ping Wang and Ken Castleman, Automated Registration of Multi-Color Fluorescence In Situ Hybridization (M-FISH) Images for Improving Color Karyotyping, Cytometry, Part A, 64A(2), April, 2005.
Given our experiences in developing industrial image processing software under SBIR research, we plan to establish a close collaboration with the industry in the next few years so that some of our research results can be commercialized.
3.Biomedical image processing
The PI, Dr. Wang, started his career on medical imaging when working as a postdoc at Washington University Medical Center for cardiovascular imaging analysis with tagged MRI. He developed a technique to analyze the movement and deformation of the human heart for clinical diagnosis. Since then, we have made contributions to various biomedical imaging such as optical imaging, microscopic imaging and fMRI imaging, ranging from tissue and organ levels to cellular and molecular levels, as evidenced from my publications.
a. Yu-Ping Wang, Y. Chen, A. A. Amini, Efficient LV Motion Estimation using Subspace Approximation Techniques, IEEE Trans. Medical Imaging, vol. 20, no.6, June 2001
b. Yu-Ping Wang, Y. Wang and P. Spencer, Fuzzy Clustering of Raman Spectral Imaging Data with a Wavelet-Based Noise Reduction Approach, Applied Spectroscopy, 60(7), 60(7), 2006.
c. Yu-Ping Wang, Husain Ragib, and Chi-Ming Huang, A wavelet approach for the identification of axonal synaptic varicosities from microscope images, IEEE Trans. Information Technology in Biomedicine, May, 11(7): 296-304, 2007.
d. Hongbao Cao and Yu-Ping Wang, Segmentation of M-FISH images for improved classification of
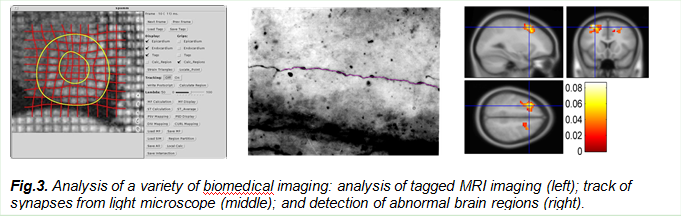
We recently received an NSF EPSCoR project with New Mexico, Nebraska, and Louisiana to form a consortium on developmental chronnecto-genomics (Dev-CoG). The overarching goal is to advance understanding of childhood brain connectivity by developing new analytic approaches to study connectivity over brief and extended periods of time (the chronnectome), via multiple neuroimaging modalities. We will continue to develop multivariate approaches for multi-scale longitudinal modeling of developmental changes in multi-modal chronnectomic measures.
4.Genomics Informatics or genomic signal processing
Genomic data analysis in the context of post-genome era information processing is an emerging and exciting topic. In the last couple years, high resolution genetic probes have evolved from Human Genome Sequencing Project, which can detect subtle and cryptic genetic aberrations unobtainable with conventional probes, thus, holding great promise for personalized medicine. Among various projects we worked, we have made contributions to the detection of copy number variations (CNVs) first from microarray platform and currently from next generation sequencing (NGS) platform. Recently, we proposed a novel statistical approach that can detect CNVs with better accuracy and more robustness. Our work has been featured in Genetic Engineering & Biotechnology News and Biocompare report.
a. Chen J, Wang YP. A statistical change point model approach for the detection of DNA copy number variations in array CGH data. IEEE/ACM Trans Comput Biol Bioinform. 2009 Oct-Dec;6(4):529-41. PubMed PMID: 19875853; PubMed Central PMCID: PMC4154476.
b. Y.-P.Wang, M. Gunampally, J. Chen, D. Bittel, M. Butler and W.-W. Cai, A Comparison of Fuzzy Clustering Approaches for Quantification of Microarray Gene Expression, Journal of VLSI Signal Processing Special Issue on Machine Learning for Microarray and Sequence Analysis, 50: 305-320, 2008.
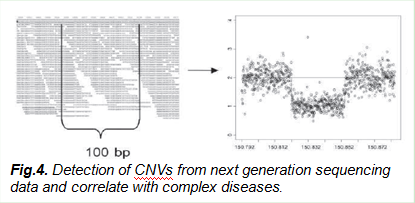
c. Duan J, Zhang JG, Deng HW, Wang YP. CNV-TV: a robust method to discover copy number variation from short sequencing reads. BMC Bioinformatics. 2013 May 2;14:150. PubMed PMID: 23634703; PubMed Central PMCID: PMC3679874.
d. Duan J, Deng HW, Wang YP. Common copy number variation detection from multiple sequenced samples. IEEE Trans Biomed Eng. 2014 Mar;61(3):928-37. PubMed Central PMCID: PMC4165854.
We are currently focusing on two projects: a). Integration of multi-omics data; and b) the analysis of next generation sequencing data. Given the availability of vast amounts of data generated by a variety of high throughput genomic techniques, how to fuse the heterogeneous multimodal genomic information to improve the predication of cancer and the response to therapies is of tremendous significance in biomedicine. We recently received a major NIH R01 award to continue this exciting research in collaboration with Tulane Center for Bioinformatics and Genomics (CBG) on the diagnosis of osteoporosis.
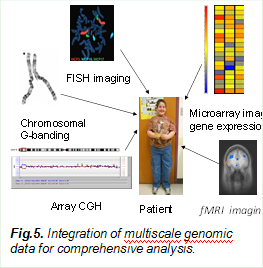
5.Imaging Genomics
In the last few years, the combination of imaging and genetic approaches has become an emerging area in brain research, where multiple complementary data are utilized for systematic and comprehensive analysis of a patient. Our current effort is on multiscale and multimodal image and genomic data integration. We pioneered the use of sparse models based approaches for data integration, which have been supported by several NIH and NSF grants. We developed a sparse group CCA model, collaborative low rank regression model and sparse regressions for the correlation and integration of (epi)-genomic and multiple imaging data, and further link with biological knowledge bases for the diagnosis of complex diseases and detection of genes.
a. Sheng J, Deng HW, Calhoun VD, Wang YP. Integrated analysis of gene expression and copy number data on gene shaving using independent component analysis. IEEE/ACM Trans Comput Biol Bioinform. 2011 Nov-Dec;8(6):1568-79. PubMed PMID: 21519112; PubMed Central PMCID: PMC3146966.
b. Cao H, Duan J, Lin D, Calhoun V, Wang YP. Integrating fMRI and SNP data for biomarker identification for schizophrenia with a sparse representation based variable selection method.
BMC Med Genomics. 2013;6 Suppl 3:S2. PubMed PMID: 24565219; PubMed Central PMCID: PMC3980348.
c. Cao H, Duan J, Lin D, Shugart YY, Calhoun V, Wang YP. Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs. Neuroimage. 2014 Nov 15;102 Pt 1:220-8. PubMed PMID: 24530838; PubMed Central PMCID: PMC4130811.
d.Yu-Ping Wang, Multiscale genomic imaging informatics, IEEE Signal Processing Magazine, Nov. Dec issue, pp. 169-172, 2009.
We currently have two ongoing R01 projects in collaboration with Prof. Vince Calhoun of Mind Research Network in Albuquerque. The project is about the integration of these multiscale and multimodal approaches, which has been promising for complex disease diagnosis and prognosis.
6.Developmental Chronnecto-Genomics
Please check the following link: Developmental Chronnecto-Genomics

