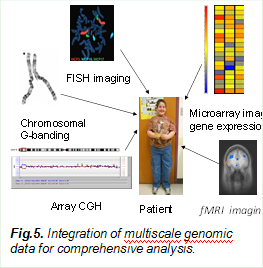
5.Imaging Genomics
In the last few years, the combination of imaging and genetic approaches has become an emerging area in brain research, where multiple complementary data are utilized for systematic and comprehensive analysis of a patient. Our current effort is on multiscale and multimodal image and genomic data integration. We pioneered the use of sparse models based approaches for data integration, which have been supported by several NIH and NSF grants. We developed a sparse group CCA model, collaborative low rank regression model and sparse regressions for the correlation and integration of (epi)-genomic and multiple imaging data, and further link with biological knowledge bases for the diagnosis of complex diseases and detection of genes.
a. Sheng J, Deng HW, Calhoun VD, Wang YP. Integrated analysis of gene expression and copy number data on gene shaving using independent component analysis. IEEE/ACM Trans Comput Biol Bioinform. 2011 Nov-Dec;8(6):1568-79. PubMed PMID: 21519112; PubMed Central PMCID: PMC3146966.
b. Cao H, Duan J, Lin D, Calhoun V, Wang YP. Integrating fMRI and SNP data for biomarker identification for schizophrenia with a sparse representation based variable selection method.
BMC Med Genomics. 2013;6 Suppl 3:S2. PubMed PMID: 24565219; PubMed Central PMCID: PMC3980348.
c. Cao H, Duan J, Lin D, Shugart YY, Calhoun V, Wang YP. Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs. Neuroimage. 2014 Nov 15;102 Pt 1:220-8. PubMed PMID: 24530838; PubMed Central PMCID: PMC4130811.
d.Yu-Ping Wang, Multiscale genomic imaging informatics, IEEE Signal Processing Magazine, Nov. Dec issue, pp. 169-172, 2009.
We currently have two ongoing R01 projects in collaboration with Prof. Vince Calhoun of Mind Research Network in Albuquerque. The project is about the integration of these multiscale and multimodal approaches, which has been promising for complex disease diagnosis and prognosis.

